Vina Hardware Acceleration
What is docking?
The goal of ligand—protein docking is to predict the predominant binding mode(s) of a ligand with a protein of known three-dimensional structure.
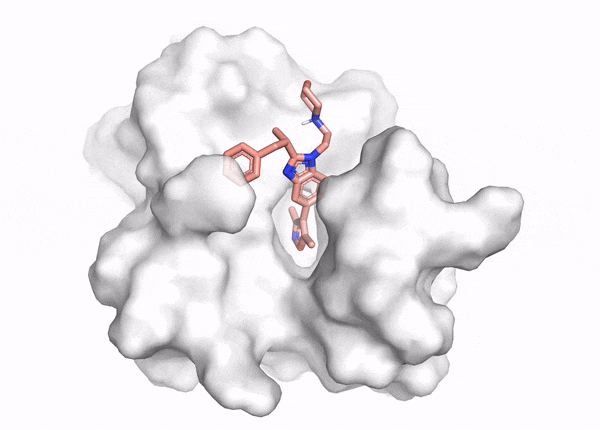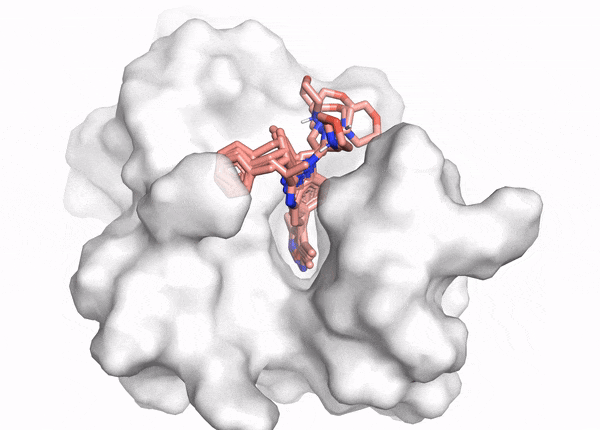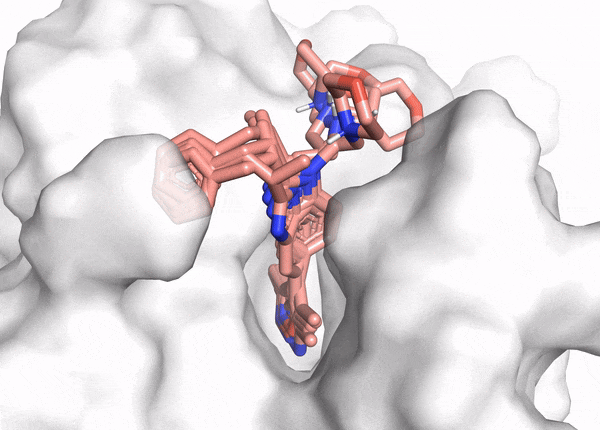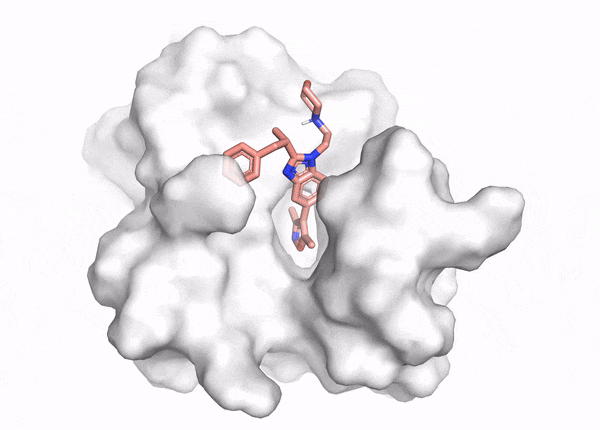
Docking demo
Why Vina?
AutoDock Vina is usually recommended as the first-line tool in the implementation of molecular docking due to its docking speed and accuracy
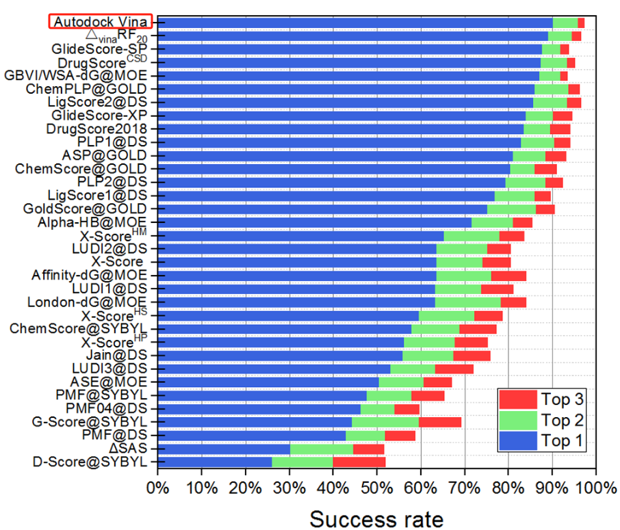
The rank of molecular docking algorithms
Vina is painfully time-consuming due to the massive combinatorial possibilities of multi-dimensional data. Previous research has managed to reduce the inference time from 475 years to 2 weeks by leveraging 10,000 CPUs.
The efforts we do for accelerating Vina
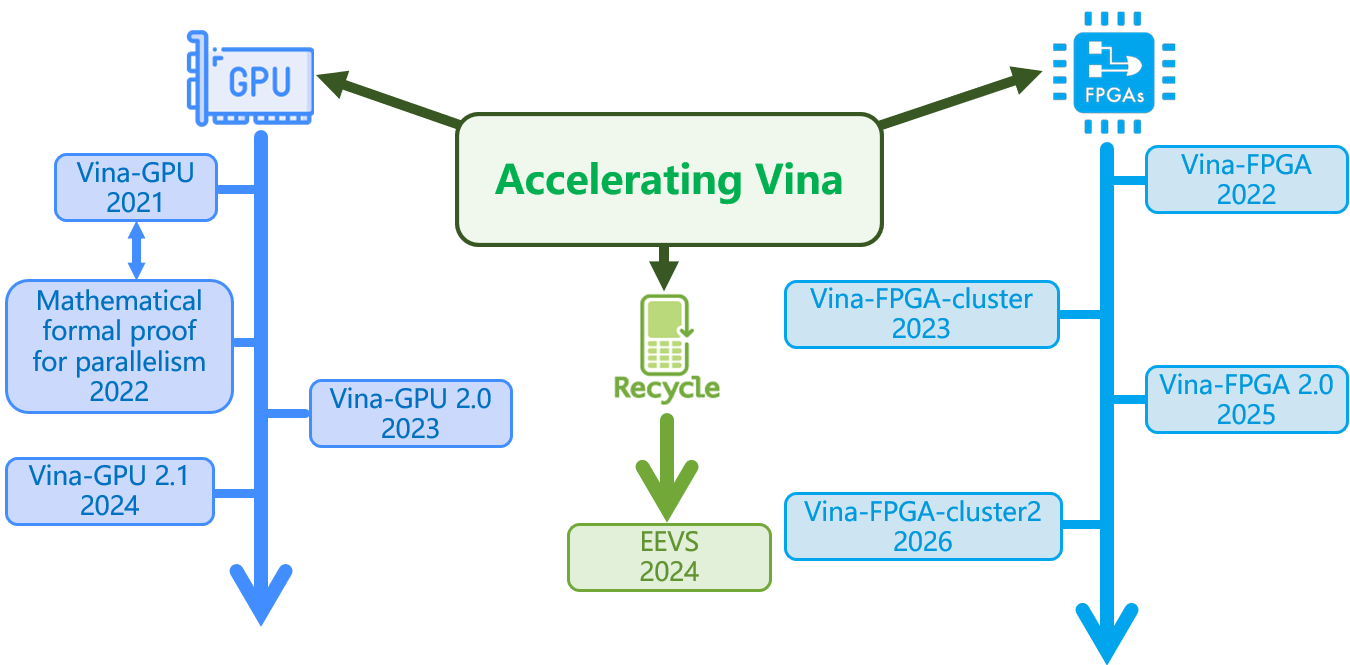
Roadmap
Vina-FPGA
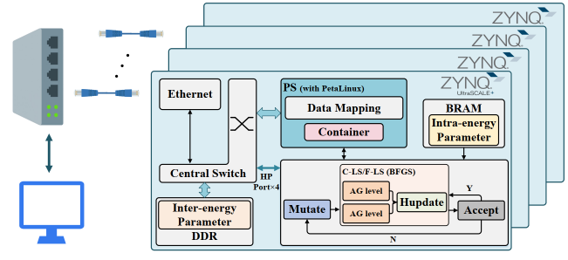
Highlights
- 1st FPGA/FPGA cluster accelerator with a significant speedup for AutoDock Vina with fixed-point quantization and low-level parallelism
- Hybrid fixed-point quantization
- Parallel calculation of inter-molecular energy and intra-molecular energy with their internal pipelines
- A novel parallel AG module design
Performance Metrics
| Metrics | CPU | Vina-FPGA | Vina-FPGA2 | Vina-FPGA-Cluster | Vina-FPGA-Cluster2 |
|---|---|---|---|---|---|
| Latency | 182.28s | 48.40s | 14.50s | 6.67s | 6.08s |
| Speed up | 1× | 3.7× | 12.6× | 27.33× | 29.9× |
| Power | 47.34W | 4.70W | 4.70W | 19.48W | 14.34W |
Publications
- Breaking the BRAM Wall: Scalable Vina FPGA Acceleration via Distributed Grid Storage and Cross-Board Long-Ring Pipelines (Vina-FPGA-Cluster2)
DATE, 2026 - Vina-FPGA 2.0: A High-Level Parallelized Hardware-Accelerated Molecular Docking Tool Based on Inter-Module Pipeline
FITEE, 2025 - Vina-FPGA-Cluster: Multi-FPGA Based Molecular Docking Tool with High-Accuracy and Multi-Level Parallelism
TBioCAS, 2024 - Vina-FPGA: A Hardware-Accelerated Molecular Docking Tool With Fixed-Point Quantization and Low-Level Parallelism
TVLSI, 2023
Vina-GPU
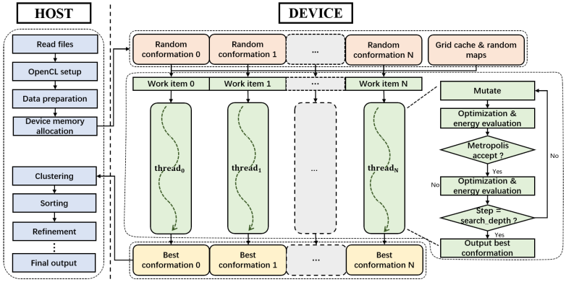
Highlights
- 1st OpenCL-based GPU implementation for Vina
- Multi-thread width control: reduce search depth without missing favorable poses
- Considerable accuracy and speedup ratio (191.68×)
Publications
- Accelerating AutoDock Vina with GPUs
Molecules, 2022 - Effectiveness Analysis of Multiple Initial States Simulated Annealing Algorithm, a Case Study on the Molecular Docking Tool AutoDock Vina
TCBB, 2023 - Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units
JCIM, 2023 - Vina-GPU 2.1: Towards Further Optimizing Docking Speed and Precision of AutoDock Vina and Its Derivatives
TCBB, 2024
Vina-Phone
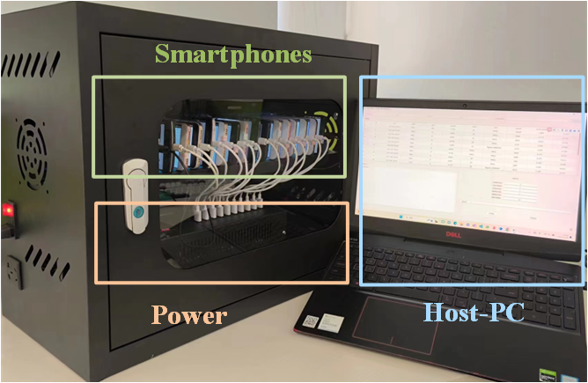
Highlights
- Leveraging the computational resources of discarded smartphones
- Cost-effective method to lower the barrier of drug discovery
- PCSA algorithm ensures thermal dissipation requirements while accelerating virtual screening